Transcriptional control of ribosomal genes is a key regulatory element in the reaction of cells to environmental cues. We study the spatial arrangement of the different epigenetically defined populations of ribosomal genes, transcriptionally active, silent and primed ribosomal genes in relation to the structural correlate of ribosomal gene expression, the nucleolus. These studies are conducted under conditions favorable for ribosome production and cell growth as well as under nutrition deprivation and treatment with inhibitors of nucleolar activity.
We employ a predominately microscopic approach including time-lapse microscopy of dCas-tagged ribosomal genes and correlated light- and electron microscopy (CLEM).
We have the exciting opportunity to study the entire lifespan of an organism by exploiting a very short-lived vertebrate aging model system, the African turquoise killifish (Nothobranchius furzeri) that is maintained in house (see Group Pusch). Specifically, we study the tissue dynamics of the killifish gut tube throughout its lifetime in order to gain insight into the inverse relationship between declining regenerative capacity and increasing cellular senescence during aging. We apply drugs that selectively reduce senescent cells (senolytics), and study the influence of dietary regimens on the regeneration of the killifish gastrointestinal tract. In relation with project 1, we investigate changes in the ribosome production and thus nucleolar function throughout the lifespan.
We employ a wide variety of biochemical, molecular, bioinformatics, and microscopic techniques.
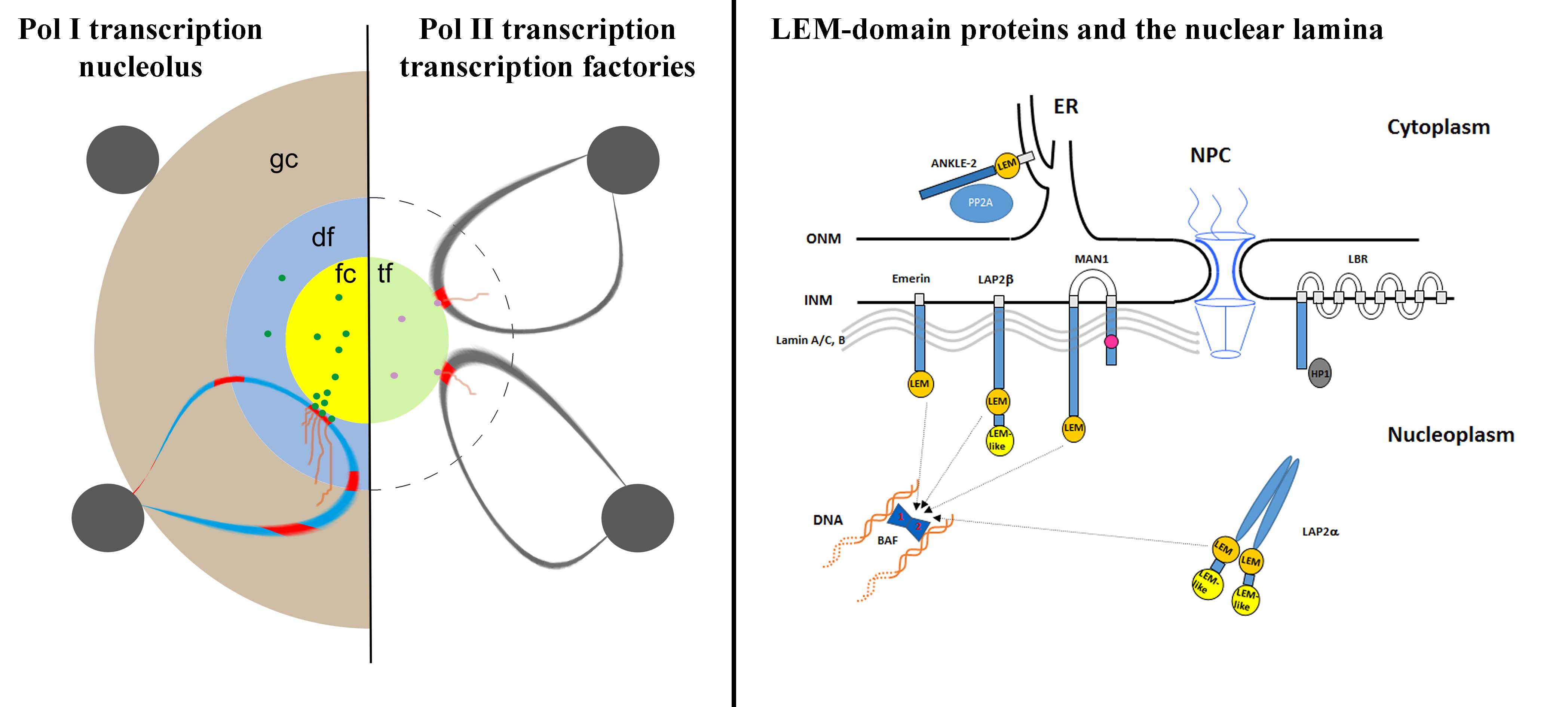
Several proteins of the inner nuclear membrane (INM) belong to the LEM-domain (Lap2-Emerin-MAN1) protein family. The LEM-domain binds BAF, which itself interacts with DNA. LEM-domain proteins have developmental functions, as exemplified by mutations in emerin causing Emery-Dreifuss Muscular Dystrophy. They also interact with transcriptional repressors. In order to examine the collective influence of the LEM-domain on chromatin architecture and post-mitotic nuclear envelope formation we are creating cell lines deficient for one or several LEM-domain proteins of the INM using the CRISPR/Cas methodology and time-lapse microscopy. The nuclear lamina studies were pioneered by Luc Snyers and discontinued after his retirement.
