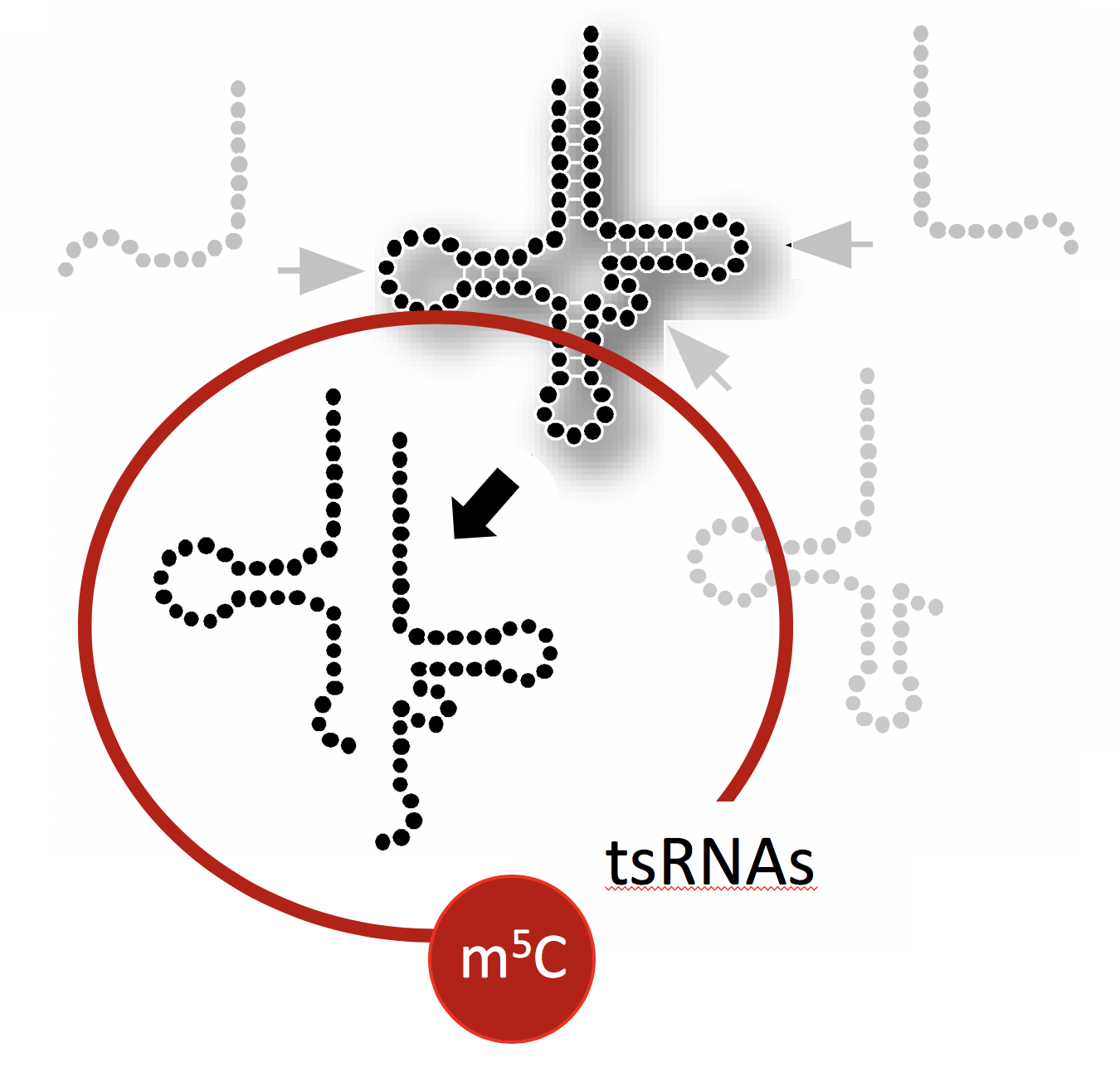
This project aims at determining the structure, protein association and physical impact of specific tsRNAs whose biogenesis is affected by the RNA modification 5-methylcytosine (m5C) mediated by the enigmatic RNA methyltransferase DNMT2/TRDMT1. To do so, biochemical and molecular biology approaches are employed to determine the structures of stress-induced tsRNAs in vivo using SHAPE-technology, to identify associated proteins, and develop reporters that inform on fhe impact of tsRNA function.
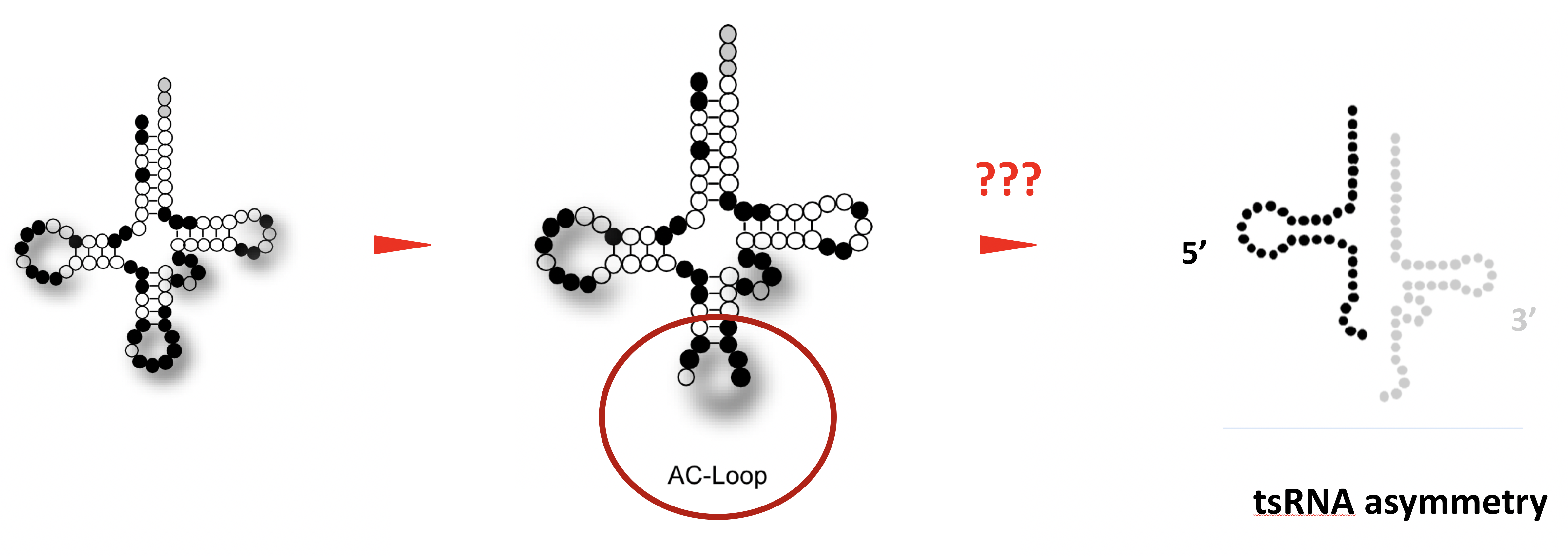
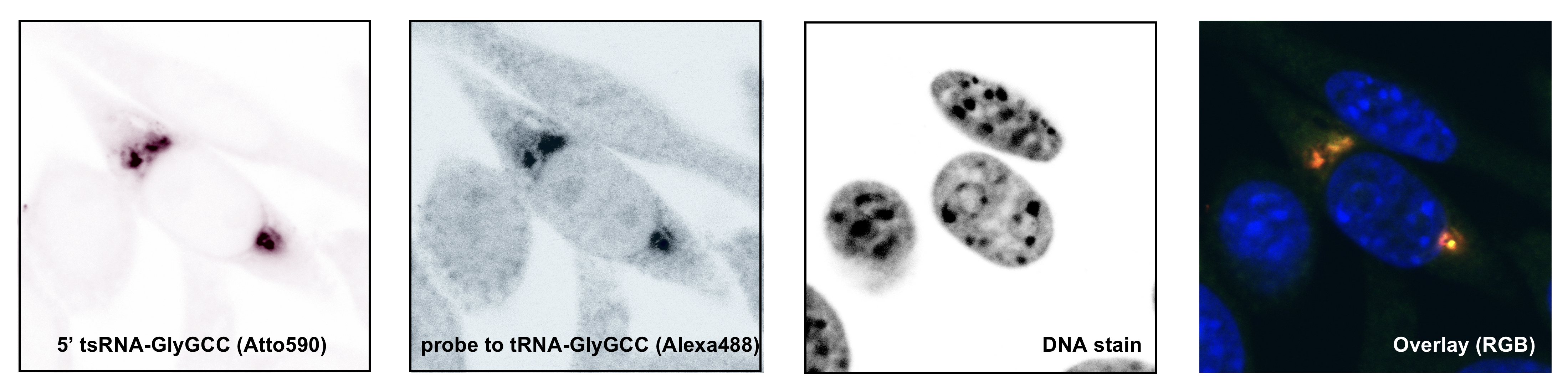
It is generally assumed that tRNA fragments are produced by the activity of enzymes, which introduce “breaks” into tRNAs. A single “break” in a tRNA should result in two tRNA fragments in equal numbers. Interestingly though, cells recovering from stress largely maintain only 5' tRNA halves, which indicates the presence of cellular activities that degrade the 3' tRNA halves.
This project will identify the enzymatic activities, which act on tRNAs that were “broken” during the stress response of human cells. Specifically, the project aims at identifying enzymes, which degrade only one tRNA half in “broken” tRNAs while leaving the other intact. To do so, the project will employ biochemical fractionation of protein complexes induced by experimental stress conditions in human cell culture systems. Once identified, the molecular details defining these activities will be determined by measuring enzymatic reaction parameters on “broken” tRNAs using recombinant protein and RNA approaches.
All experimental manipulations using tsRNAs were performed by injecting or transfecting relatively large copy numbers of tsRNAs or tsRNA-containing RNA preparations into cells. This raises the question as to whether the reported phenotypes were caused by small RNAs reflecting physiological relevant copy numbers, or could have been the result of RNA-mediated mass effects. To address these questions, we aim at measuring absolute copy numbers of various tsRNAs, which will allow for a better quantitative understanding of the biological system that is being manipulated with ectopic tsRNAs.